Research
Tightly linked to the Boston Medical Center (BMC), we ask ourselves fundamental questions such as: How can we improve the quality of patients’ lives? What drives and controls cellular plasticity in development and tumor resistance? How do cells transcriptionally respond to targeted therapies and how can we steer it in the right direction? What is the role of signaling and cell communcation within the (tumor) microenvironment? And how can we ultimately leverage our knowledge to identify better biomarkers or novel therapy strategies?
To answer these questions we make use of state-of-the-art facilities and emerging spatial technologies and apply them directly on primary tumor tissues. Alternatively, we aim to mimick such biological processes using 3D multi-cellular in vitro models, such as (tumor) organoids and spheroids. These systems can then be challenged through genetic engineering and be profiled using a wide-array of genomics and imaging technologies.
We focus on biological processes that control the decision making of an individual (cancer) cell. These processes range from extracelluar cues, such as signaling pathways and cell-to-cell interactions, to intracellular mechanisms that control the cell’s fate and its ability to (de-)differentiate, such as chromatin reorganization and transcriptional processing.
Current Projects
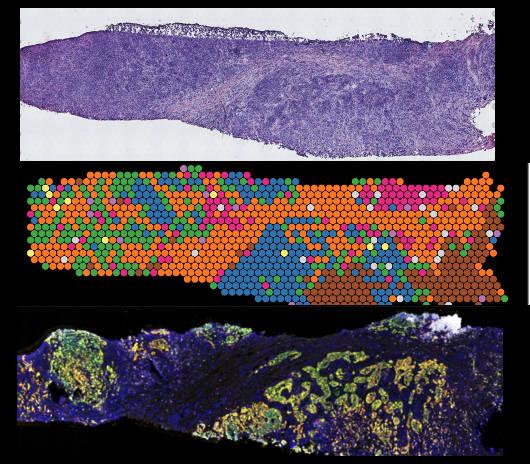
Cancer Disparities in Triple Negative Breast Cancer
Triple negative breast cancer is the most aggressive breast cancer and is frequently seen in Black Women. Here we seek to identify the spatial topological configurations and drivers that promote the aggressive nature of this tumor type. By utilizing a set of tumor samples from a large and diverse patient population we aim to improve prediction to treatment strategies and to identify novel vulnerabilities that work in all populations. In this manner we are part of a larger and enthusiastic collaboration that aims to eliminate cancer disparities.
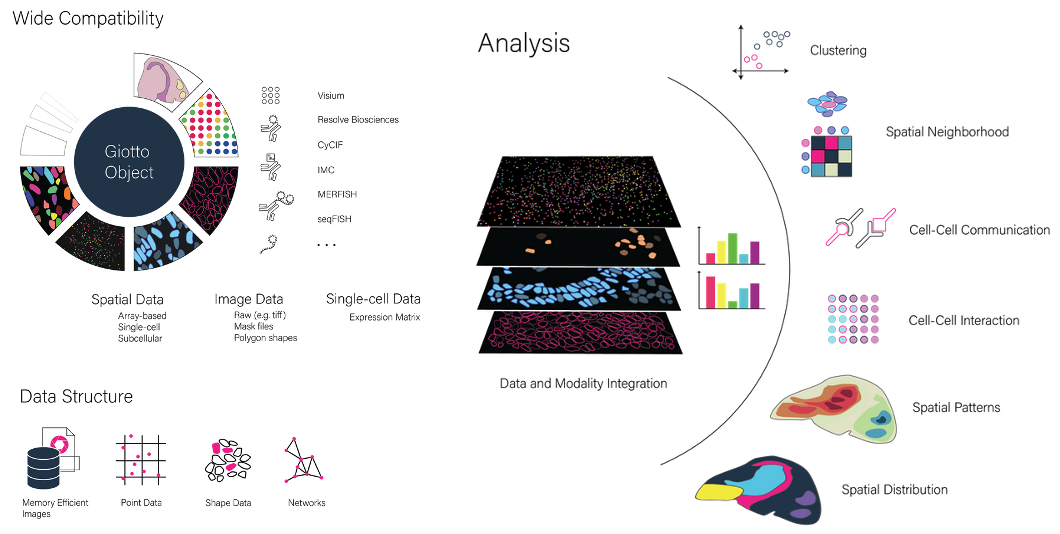
Multi-resolution and multi-modal data structures for integrative spatial omics analysis
Spatial (multi-)omics is an emerging field that allows researchers to interrogate the importance of spatial location and cellular cross-talk in their favorite tissue, model organism or biological processes. Different spatial technologies provide different sources of information and they often do this at different scales. We continue building on top of our Giotto expertise and are working on creating harmonized data structures that allow processing, integrating and jointly analyzing virtually all spatial datasets. We specifically focus on developing tools in an open-source setting and that can be integrated with other existing and established tools, frameworks or communities.

Pulmonary Metastasis in Osteosarcoma
Pediatric osteosarcoma, a tumor that affects children during growth spurts, is a challenging disease to treat, and treatment has not improved in almost four decades. The primary cause of mortality for pediatric osteosarcoma is metastases to the lung. This interdisciplinary project, funded by Alex’s Lemonade Stand, seeks to uncover the processes that drive the development of metastatic lesions. Using cutting edge technology including spatial transcriptomics, we hope to identify druggable drivers of metastasis.
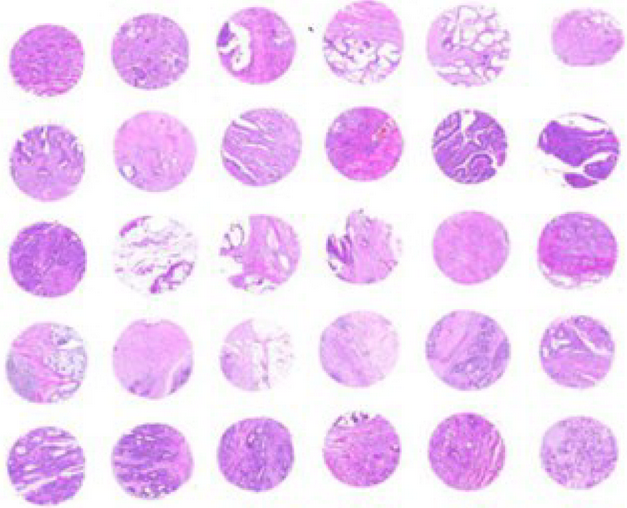
Neuroendocrine tumor heterogeneity
Neuroendocrine tumors display exceptional heterogeneity that underlie their resistance to standard and novel treatment strategies. Here we will use several cutting-edge spatial technologies and data analysis strategies to decode the intrinsic and extrinsic factors that contribute to this observed complexity. We aim to translate our insights into the development of improved biomarkers or the identification of new targettable molecules within the whole tumor microenvironment.
