Publications
2024
Clinical Cancer Research · 14 Nov 2024
Genome Biology · 01 Aug 2024
2023
Cold Spring Harbor Laboratory · 27 Nov 2023
2022
Frontiers in Endocrinology · 01 Dec 2022
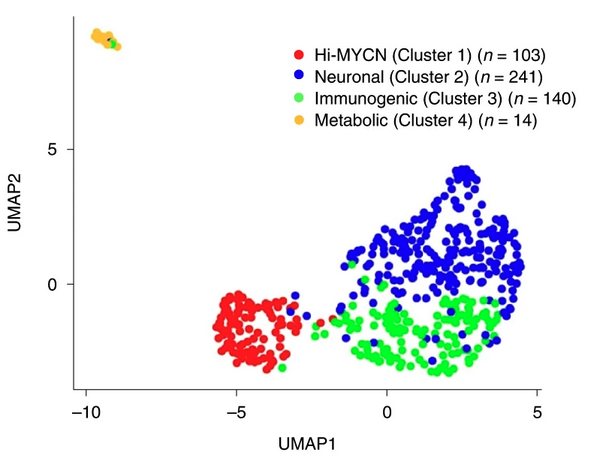
Nature Cancer · 22 Sep 2022
Current Protocols · 01 Apr 2022
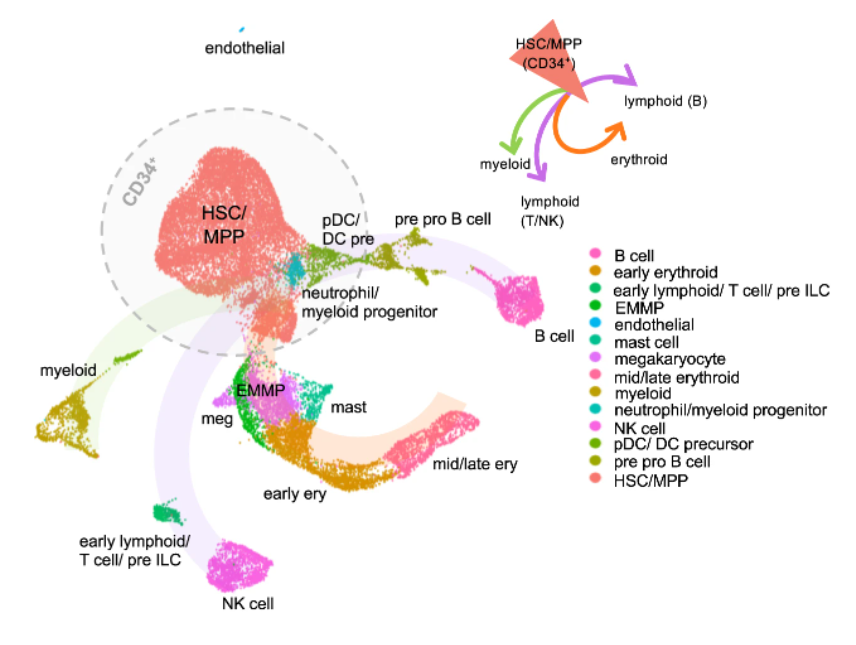
Nature Communications · 01 Mar 2022
2021
Genome Research · 01 Oct 2021
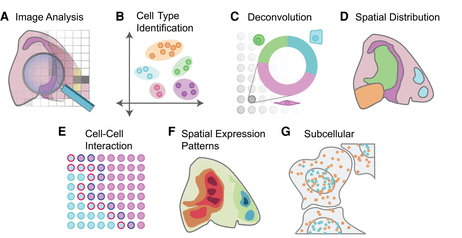
Genome Biology · 08 Mar 2021
2020
Cancer Cell · 01 Jan 2020
2019
Nature · 01 Aug 2019
Nature Communications · 15 Apr 2019
Nature · 25 Mar 2019